doi: 10.3389/fviro.2022.834808
Among numerous point mutation differences between the SARS-CoV-2 and the bat
RaTG13 coronavirus, only the 12-nucleotide furin cleavage site (FCS) exceeds 3
nucleotides. A BLAST search revealed that a 19 nucleotide portion of the SARS.Cov2
genome encompassing the furing cleavage site is a 100% complementary match to a
codon-optimized proprietary sequence that is the reverse complement of the human
mutS homolog (MSH3). The reverse complement sequence present in SARS-CoV-2
may occur randomly but other possibilities must be considered. Recombination in an
intermediate host is an unlikely explanation. Single stranded RNA viruses such as SARS-
CoV-2 utilize negative strand RNA templates in infected cells, which might lead through
copy choice recombination with a negative sense SARS-CoV-2 RNA to the integration
of the MSH3 negative strand, including the FCS, into the viral genome. In any case,
the presence of the 19-nucleotide long RNA sequence including the FCS with 100%
identity to the reverse complement of the MSH3 mRNA is highly unusual and requires
further investigations.
Keywords: SARS-CoV-2 spike, furin cleavage site, MSH3 gene, sequence homology, recombinability
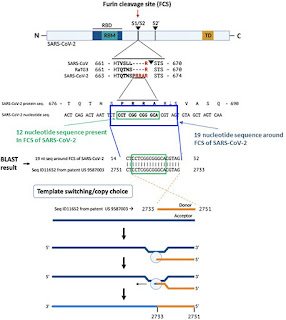
FIGURE 1 | The origin of the furin sequence in SARS-CoV-2. Comparison of the protein sequences at the S1/S2 junction in SARS-CoV, RaTG13, and SARS-CoV-2
demonstrating the presence of the furin cleavage site (FCS) PRRA only in SARS-CoV-2. Based on a BLAST search of the 12-nucleotide stretch coding for the FCS
PRRA, a 19-nucleotide long identical sequence was identified in the patented (US 958 7003) sequence Seq ID11652. SEQ ID11652 is transcribed to a MSH3 mRNA
that appears to be codon optimized for humans. This 19-nucleotide sequence including 12 nucleotides coding for the FCS PRRA, present in the human MSH3 gene
might have been introduced into the SARS-CoV-2 genome by the illustrated copy choice recombination mechanism in SARS-CoV-2 infected human cells
overexpressing the MSH3 gene.

No comments:
Post a Comment